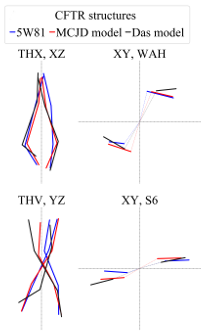
Since an increasing number of ABC membrane protein structures are determined by cryo-electron microscopy, identifying differences between their conformations has become an arising issue. Therefore, we propose to define standardized measures for ABC structure characterization. We set conformational vectors, conftors, which describe the relative orientation of domains and can highlight structural differences. ABC conftors have been defined between e.g. the ends of TM helices, the intracellular end of specific TM helices and the center of masses (COM) of coupling helices, and the COM of coupling helices and a specific region (S10) in the nucleotide binding domain (top right figure). Conftors can be used for various purposes, such as describe differences in conformations associated to functional states and analyze conformational changes in MD simulations. In addition, conftors can be projected to 2D for highlighting structural differences, as shown in the figure on the bottom right, including various CFTR structural models.
Conftors can be applied not only to the ABC superfamily. However, for other classes of proteins several vectors should be tested by an expert of the given protein family as long as no automatic algorithms are available. This web application is the very first step of generalization of standardized geometric measures for structure comparison. Currently a basic set of conftors based on TM helices and membrane normal are calculated and custom conftors based on these entities can be defined and calculated as well. In addition, the target structures are limited to those, which are present in the PDBTM database, since TM helix definition is taken from this data source. Our plan is to develop algorithms for automatic definition and testing of conftors for each membrane protein class.
Please provide feedback on bugs and any requested feature!
Reseach support:
Please visit our sites: